News - 2025-10-28
Hello everyone! This newsletter is a few weeks later than anticipated, so sorry for the delay. It’s so nice to see the new faces in our HPC community and to welcome back our returning users.
This month’s newsletter details:
New NVIDIA H100 NVL GPU nodes are available on Stanage.
Community Software Areas.
Matlab Updates.
New Matlab container.
Changes to the maximum number of active GPU jobs allowed per user.
Reminder notice of Bessemer decommissioning (just a few days to go!).
New software installations.
Lustre filesystem usage warning.
Upcoming training courses.
New H100 NVL GPU Nodes on Stanage
We are excited to announce that we have added two new H100 GPU nodes to Stanage in the gpu-h100-nvl partition. Each node is equipped with 4 new NVIDIA H100 NVL GPUs each for a total of eight new GPUs. These GPUs are designed for high-performance computing and AI workloads, delivering significant performance improvements over the older NVIDIA H100 PCIe GPUs including:
Increased Performance: The H100 NVL GPUs provide enhanced computational power, with 15% more CUDA and tensor cores.
Larger Memory Capacity: The NVL variant comes with 94 GB memory, an 18% increase on existing H100 nodes, allowing for larger datasets and models.
Increased Memory Bandwidth: The H100 NVL GPUs provide 95% more memory bandwidth than the older H100 PCIe GPUs
The full specifications of the new H100 NVL nodes are detailed here. From our initial testing, we have seen performance improvements of up to 1.51x on our pytorch benchmarks compared to the older H100 PCIe nodes. These tests were run using different ResNet layers and batch sizes. Below are some benchmark results showing how many seconds it took to complete the tests on both the H100 PCIe and H100 NVL nodes:
Cuda |
ResNet Layers |
Batch Size |
h100 |
h100-nvl |
Speed up |
12.1 |
18 |
32 |
30.1466 |
25.8455 |
1.17x |
12.1 |
18 |
128 |
26.7622 |
24.237 |
1.1x |
12.1 |
50 |
32 |
59.7228 |
44.8023 |
1.333x |
12.1 |
50 |
128 |
55.3886 |
40.4741 |
1.35x |
12.1 |
152 |
32 |
120.64 |
85.1803 |
1.4x |
12.1 |
152 |
128 |
115.24 |
76.2963 |
1.51x |
These results are visualised in the plot below:
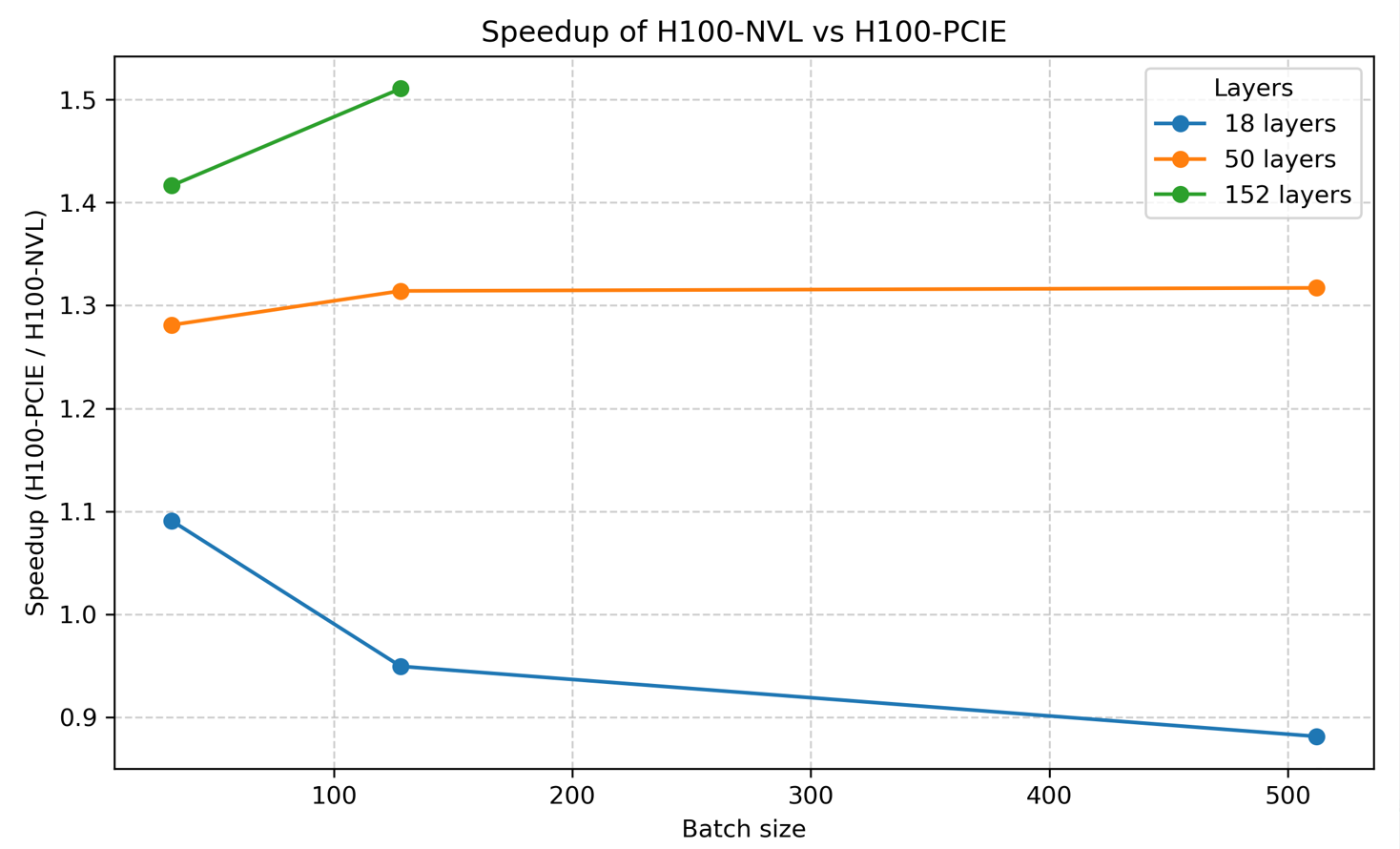
The increased memory capacity also allowed us to run larger models on the H100 NVL nodes that were previously impossible. For example, with ResNet152 we were able to run at full batch size of 512 on the H100 NVL GPUs compared to a maximum of 128 on the older H100 PCIe GPUs. We cannot wait to see how our users will take advantage of these new nodes for their research workloads! The documentation for using the H100 NVL nodes is available here.
Note
The A100 and older H100 PCIe nodes have AMD CPUs (znver), while the new H100 NVL nodes have Intel CPUs (icelake). In some cases your software may need to be re-compiled depending on which GPU node it is to be run on.
Community Software Areas
In response to user feedback on the need for an area on Stanage for storing research-group managed software/scripts, we are pleased to introduce Community Software Areas to Stanage. This storage space is for hosting common software installations for your research group or broader Sheffield HPC community. Details on how to request, manage and utilise these areas are detailed in our Community Software Areas documentation.
Matlab Updates
Over the last few years our advice on how to use MATLAB non-interactively has been to use the following command in batch scripts.
matlab -nosplash -nodesktop -r myscript
However, since R2019a, MATLAB has had a -batch command line switch and we now recommend that you use the following instead:
matlab -nodisplay -batch myscript
The benefits of using -batch are well documented on the Mathworks blog (written by RSE Sheffield alumnus Mike Croucher) but in summary -batch:
Runs in true non-interactive mode — no desktop, no splash screen, no input prompts. The previous command could still prompt for input in some cases
Is less noisy at the beginning of execution, reducing clutter in logs.
Automatically exits MATLAB when the script or function finishes executing or if an error occurs. The exit code returned to the OS reflects success (0) or failure (non-zero), which is critical for CI/CD pipelines or shell scripts.
Supports better error handling and reporting.
Matlab Container Update
Due to Stanage’s older operating system (EL7), some newer versions of software cannot be installed due to OS-level dependencies. We have plans to update the operating system to EL9, but in the interim to address this incompatibility we have been providing container images for several applications. Among these, we recently created a new MATLAB container image for MATLAB R2025a thanks to one of the members of our HPC community requesting it. This container image includes support for GPU computing with the Parallel Computing Toolbox and is optimised for use on our A100 and H100 GPU nodes. One important thing to be aware is that with it being container-based it cannot (easily) be used within multi-node jobs.
You can load the new container with the following command:
module load Matlab/2025aContainer
We are still running tests on this container with the user who requested it and should be publishing the documentation for it soon, but for those who want to experiment please don’t hesitate to reach out to us and we will send you a guide on how to use it.
Changes To Maximum Number Of Active GPU Jobs
Due to the increased demand for GPU resources on Stanage and feedback from our HPC community, we have adjusted the maximum number of concurrent GPUs per user. The new limit is 12 active GPUs (previously 16). This change is intended to balance the growing needs of our researchers with fair access for all, reducing the risk of any single person using the majority of the available shared A100 and H100 nodes.
Our usage policies are regularly reviewed and updated to ensure they meet the needs of our users. We would like to thank those of you who provided feedback and would love to hear your view on this change or any other aspect of our service. If you would like to share anything that you feel would improve the overall user experience on the Stanage HPC, please do not hesitate to get in touch via mailto:research-it@sheffield.ac.uk.
Bessemer Decommissioning
Our Bessemer cluster is planned to be decommissioned on Friday 31st October 2025. After more than 6 years of service it is time to retire the system. To prepare, we ask all users to:
Transfer all relevant/needed data/files/models (in
/home&/fastdata) to Stanage. Bessemer/home&/fastdataareas will become inaccessible post 1700hrs 31st October 2025Confirm that the software you currently use on Bessemer is available on Stanage (do not assume it is) - if it’s not then you need to advise us as soon as possible so we can look into getting it installed on Stanage. You can check if the software available on Stanage by running
module spider <SOFTWARE_NAME>on Stanage or by looking at the HPC documentation . If you need new software installed, please raise a ticket via mailto:research-it@sheffield.ac.uk.Test your workloads on Stanage
Request your Research Storage (
/shared) to be mounted on Stanage (if you have not already done so). You will also need to confirm that your/sharedareas do not have any sensitive information stored. Please raise a ticket to request this & confirm that you do not have sensitive data stored there
Warning
Shared Research Storage areas are only available on Stanage login nodes and NOT worker nodes as they have been on Bessemer. You will need to amend your workflows to take this change into account.
In the meantime, please do not hesitate to reach out if you have any questions or concerns. Remember, the earlier you migrate to Stanage, the less stressful it will be to you and the HPC support team in October.
New software installations
We have recently installed the following new software on Stanage:
Nextflow/25.04.6: Nextflow is a reactive workflow framework and a programming DSL that eases writing computational pipelines with complex data. Nextflow documentation.
SimNIBS/4.0.1-foss-2023a: SimNIBS is a free and open source software package for the Simulation of Non-invasive Brain Stimulation. (Documentation Pending)
RepastHPC-Boost1.73.0/2.3.1-foss-2018b: The Repast Suite is a family of advanced, free, and open source agent-based modeling and simulation platforms that have collectively been under continuous development for over 15 years: Repast for High Performance Computing 2.3.1 is a lean and expert-focused C++-based modeling system that is designed for use on large computing clusters and supercomputers. (Documentation Pending)
Ruby/3.2.2-GCCcore-12.2.0: Ruby is a dynamic, open source programming language with a focus on simplicity and productivity. It has an elegant syntax that is natural to read and easy to write. (Documentation Pending)
Further, to accommodate our new NVIDIA H100 NVL GPUs that reside in nodes with an Intel architecture, we have updated our icelake software stack to contain the same GPU-related packages as our znver3 stack.
Lustre filesystem usage
This area is currently quite full (78% utilisation). Please can you remove any data from this filesystem that you no longer need by either deleting it or migrating what you want to e.g. a Shared Research Area. Also, please keep in mind that the Lustre filesystem in Stanage should primarily be treated as a temporary file store: it is optimised for performance and has no backups, so any data of value should not be kept on Lustre long-term.
Upcoming Training
Below are our key research computing training dates for October and the rest of this semester. You can register for these courses and more at MyDevelopment .
Warning
For our taught postgraduate users who don’t have access to MyDevelopment, please email us at mailto:researchcomputing@sheffield.ac.uk with the course you want to register for, and we should be able to help you.
29/10/2025 - Python for Data Science 1.
30/10/2025 - Introduction to Linux and Shell Scripting.
04/11/2025 - Introducing AI into Research.
05/11/2025 - Python for Data Science 2.
07/11/2025 - Python Programming 1.
12/11/2025 - Supervised Machine Learning.
14/11/2025 - Python Programming 2.
18/11/2025 - Introducing AI into Research.
19/11/2025 - Unsupervised Machine Learning.
20/11/2025 - Introduction to SQL.
21/11/2025 - Python Programming 3.
25/11/2025 - R/Python Programming 1.
26/11/2025 - Neural Networks.
28/11/2025 - Python Profiling and Optimisation.
02/12/2025 - Introducing AI into Research.
04/12/2025 - R/Python Programming 2.
09/12/2025 - HPC Training Course.
Below are some training from our third party collaborators:
EPCC, who provide the ARCHER2 HPC service, are running the following training sessions:
19/11/2025 - Parallel Python . You can register for the course here .
25/11/2025 - Efficient Parallel IO . You can join register for the session here .
Useful Links
RSE code clinics . These are fortnightly support sessions run by the RSE team and IT Services’ Research IT and support team. They are open to anyone at TUOS writing code for research to get help with programming problems and general advice on best practice.
Training and courses (You must be logged into the main university website to view).