Frequently Asked Questions
In this section, we’ll discuss some tips for solving problems encountered on Stanage. It is suggested that you work through some of the ideas here before contacting the IT Services Helpdesk for assistance.
Strange things are happening with my terminal or my terminal seems broken
Symptoms include many of the commands not working and just bash-4.1$ or sh-4.2$ being displayed instead of your username at the bash prompt.
This may be because you’ve deleted your .bashrc and .bash_profile files - these are ‘hidden’ files which live in your home directory and are used to correctly set up your shell environment.
If you hit this problem you can run the command resetenv which will restore the default files, then you should logout and log back in.
Note
If the command restenv is not working (possibly removed from $PATH), you can run the resetenv command directly:
/opt/site/bin/resetenv
I can no longer log in
If you are confident that you have no password entry issues, have already requested and been granted a HPC account and are connected to the VPN but you still can not log onto a cluster,
you may have inadvertently corrupted your shell environment if you have been installing software or making changes in your .bashrc file. Please attempt to resolve this first by resetting
your environment with the following command, replacing the variables appropriately: ssh -t $USER@$CLUSTER.shef.ac.uk 'resetenv -f'
Alternatively, you may be having problems due to exceeding your cluster filestore quota. If you exceed your filestore quota in your $HOME area it is sometimes possible that crucial
files in your home directory get truncated which effect or prevent the login process.
If the resetenv -f command does not resolve your issue and you suspect your $HOME area is full , you should contact
research-it@sheffield.ac.uk and ask to be unfrozen providing your username and a list files or folders which can be removed
or temporarily moved to your /mnt/parscratch area on Stanage.
How do I find out more about the commands available on the clusters with man pages?
Man pages (manuals) are installed by default alongside commands/software on unix-like operating systems such as those found on the clusters.
To view the man page (official manual) for a command, you can use the command:
man <command_name>
You can navigate man pages using (the same keyboard shorcuts as less):
Space to advance one page
d to advance half a page
b to go back one page
u to go back half a page
/ starts search mode, after which you enter a search term
Whilst in search mode press n for next occurrence and N for previous occurrence.
You can also press h when viewing man pages to show help.
Documentation for less is available on the system using the command:
man less
If you don’t know the specific command name then you can use apropos which searches all names of man pages and their one line descriptions:
apropos <search_term>
To list all avaiable command man pages:
apropos .
I’ve loaded software but it isn’t working
This usually means that you are on a login node. You will need to start an interactive session after which you will be able to load cluster software.
srun --pty bash -i
My batch job terminates without any messages or warnings
When a batch job is initiated by using the sbatch commands, it gets allocated specific amount of real memory and run time that you request, or small default values.
If a job exceeds either the real memory or time limits it gets terminated immediately and usually without any warning messages.
It is therefore important to estimate the amount of memory and time that is needed to run your job to completion and specify it at the time of submitting the job to the batch queue.
Please refer to our Choosing appropriate compute resources page for information on how to assess sensible resource amounts and avoid these problems.
Tip
If you are confident that the scheduler is not terminating your job, but your job is prematurely stopping, please check if you have attempted to exceed your disk space quota, instructions for this are seen below.
I’ve submitted a job but it’s not running
I submitted a job and after several days it is still waiting in the queue. How can I resolve this? There are a multitude of factors which could be causing your job to queue for a long time or to not run at all. Occasionally parts of the system may be in a maintenance period or may be utlised to capacity. A few things to consider which would cause your job to not run at all:
Did you request an acceptable amount of memory for a given node? (e.g. on Stanage, 2TB or less.)
Did you request too much time? (e.g for Stanage, more than 96 hours.)
Following are ways to fix too much time requested:
The maximum run time for Stanage is 96 hours.
You can get an estimate for when your job will run on Stanage using:
squeue --start -j <jobid>
You can reduce the runtime using:
scontrol update jobid=<job_id> TimeLimit=<new_timelimit>
then to verify the time change type:
squeue -j <jobid> --long
Alternatively, delete the job using scancel and re-submit with the new max runtime.
Case sensitivity
The High-Performance Computers at Sheffield run a Linux operating system, which is case sensitive. When logging in your username should be written like te1st and not TE1ST. Similarly, files named my_file.txt can not be opened using MY_FILE.TXT and the original matching case version should be used.
“No space left on device”, “Disk quota exceeded” errors and jobs prematurely stopping
Each user of the system has a fixed amount of disk space available in their home directory. If you see an error in your job’s logs indicating “No space left on device” or “Disk quota exceeded” it is likely that your quota has ran out.
If you attempt to exceed this quota, various problems can emerge such as an inability to launch applications or run jobs, the inability to login or abruptly terminated jobs
as programs or executables are now unable to write to your /home folder.
To see if you are attempting to exceed your disk space quota, run the quota command:
[te1st@login1 [stanage] ~]$ quota -u -s
Filesystem space quota limit grace files quota limit grace
storage:/export/users
3289M 51200M 76800M 321k* 300k 350k none
In the above, you can see that the ‘soft’ space quota is 50 gigabytes and a small portion of this is currently in use. However, the files ‘soft’ quota is 300k which has been exceeded,
additionally the grace period indicates the grace period for exceeding the soft quota has expired.
Any jobs submitted by this user will likely result in an Eqw status.
The recommended action is for the user to delete enough files, or move enough files to another filestore to allow normal work to continue.
To assess what is using up your quota within a given directory, you can make use of the ncdu module on Stanage. The ncdu utility will give you an interactive display of which files or folders are taking up storage in a given directory tree.
Conda environments are a common cause of users exceeding their storage quota.
Sometimes, it is not possible to log in to the system because of a full quota. In this situation you should contact research-it@sheffield.ac.uk
to ask for assistance, providing your username and a list files or folders which can be removed or temporarily moved to your /mnt/parscratch
area on Stanage.
I am getting warning messages and warning emails from my batch jobs about insufficient memory
If a job exceeds its real memory resource it gets terminated.
These errors on Stanage will be noted in the job record or sent via email and will resemble:
Slurm Job_id=12345678 Name=job.sh Failed, Run time 00:11:06, OUT_OF_MEMORY
To query if your job has been killed due to insufficient memory please see the cluster specific “Investigating finished jobs” sections on our Job Submission and Control page.
To request more memory and for information on how to assess sensible resource amounts please refer to our Choosing appropriate compute resources page.
Insufficient memory in an interactive session
By default, an interactive session on Stanage provides you with 4016 MB of memory.
You can request more than this when running your srun command.
$ srun --mem=8G --pty bash -i
This asks for 8 Gigabytes of RAM (real memory).
Hint
You cannot request more memory than a single node possesses and the larger the memory request, the less likely the interactive session request is to succeed. Please see the cluster specific “Interactive jobs” sections on our Job Submission and Control page.
‘Illegal Instruction’ errors
If attempts to run a binary executable program fail with an Illegal Instruction error then
your executable program (or a dynamically-linked library) may have been compiled so as to
make more optimal use of the instruction set of a particular CPU architecture (an optimised binary),
but you’re running the executable on CPU(s) that use a slightly different instruction set.
For example, you may have a executable program optimised for the Intel Icelake CPU instruction set but you find it fails to run on AMD Milan CPUs, or you might be trying to run a binary optimised for a very new Intel CPU architecture on an older model of Intel CPU.
Important
For the above reasons we recommend that you avoid copying binary executables on to the HPC systems and instead (re)compile programs and libraries on the HPC systems instead where possible.
This has the added benefits of ensuring that:
Programs/libraries are compiled against the versions of dependencies provided on the HPC systems.
Programs/libraries are more likely to make use of the more advanced features of the CPU models in the HPC systems, which could result in better performance/efficiency.
sbatch: error: Batch script contains DOS line breaks (rn) errors
If you prepare text files such as your job submission script on a Windows machine, you may find that they do not work as intended on the HPC systems.
The reason for this behaviour is that Windows and Unix machines have different conventions for specifying ‘end of line’ in text files. Windows uses the
control characters for ‘carriage return’ followed by ‘linefeed’, \r\n, whereas Unix uses just ‘linefeed’ \n.
This means a script prepared in Windows using Notepad which looks like this:
#!/bin/bash
echo 'hello world'
will look like the following to programs on a Unix system:
#!/bin/bash\r\n
echo 'hello world'\r\n
For example, if you uploaded a submission script (test.sh) with windows line endings to the cluster, then tried to submit the script using sbatch, you
would see the following:
[te1st@stanage-login1 ~]$ sbatch test.sh
sbatch: error: Batch script contains DOS line breaks (\r\n)
sbatch: error: instead of expected UNIX line breaks (\n).
If you have seen this error or suspect that this is affecting your jobs, run the following command on the file at the terminal
$ dos2unix your_files_filename
You should set your text editor to use Linux endings to avoid this issue.
error: no DISPLAY variable found with interactive job
If you receive the error message:
error: no DISPLAY variable found with interactive job
the most likely cause is that you forgot the -X switch when you logged into the cluster. That is, you might have typed:
ssh username@clustername.shef.ac.uk
instead of:
ssh -X username@clustername.shef.ac.uk
macOS users might also encounter this issue if their XQuartz is not up to date.
macOS users should also try -Y if -X is not working:
ssh -Y username@clustername.shef.ac.uk
Problems connecting with WinSCP
Some users have reported issues while connecting to the system using WinSCP, usually when working from home with a poor connection and when accessing folders with large numbers of files.
In these instances, turning off Optimize Connection Buffer Size in WinSCP can help:
In WinSCP, goto the settings for the site (ie. from the menu
Session->Sites->SiteManager)From the
Site Managerdialog click on the selected session and click edit buttonClick the advanced button
The Advanced Site Settings dialog opens.
Click on connection
Untick the box which says
Optimize Connection Buffer Size
Problems connecting with FileZilla due to MFA
Due to use of MFA (multi-factor authentication) you might find FileZilla constantly asking for your password. Two simple changes are needed to connect using FileZilla to the HPC clusters efficiently.
Change the logon type to interactive login.
Limit the number of simultaneous connections to 1.
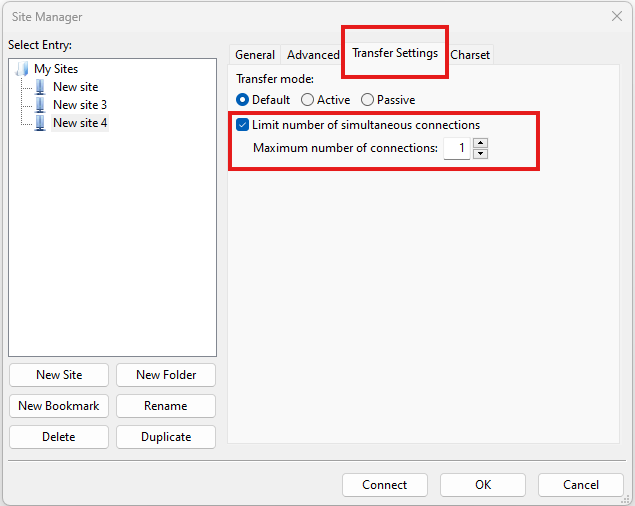
Detailed instructions are contained in the following link: https://unm-student.custhelp.com/app/answers/detail/a_id/7857/~/filezilla-ftp-configuration-for-duo-mfa-protected-linux-servers
Strange fonts or errors re missing fonts when trying to start a graphical application
Certain programs require esoteric fonts to be installed on the machine running the X server (i.e. your local machine).
Example of such programs are qmon, a graphical interface to the Grid Engine scheduling software, and the ANSYS software.
If you try to run qmon or ANSYS software on a Linux machine and see strange symbols instead of the Latin alphabet or get an error message that includes:
X Error of failed request: BadName (named color or font does not exist)
Then you should try running the following on your own machine:
for i in 75dpi 100dpi; do
sudo apt-get install xfonts-75dpi
pushd /usr/share/fonts/X11/$i/
sudo mkfontdir
popd
xset fp+ /usr/share/fonts/X11/$i
done
Warning
Note that these instructions are Ubuntu/Debian-specific; on other systems package names, paths and commands may differ.
Next, try connecting to a cluster using ssh -X clustername.shef.ac.uk, start a graphical session then try running qmon or ANSYS software again.
If you can now run qmon or ANSYS software without problems then you need to add two lines to the .xinitrc file in your home directory on your own machine
so this solution will continue to work following a reboot of your machine:
FontPath /usr/share/fonts/X11/100dpi
FontPath /usr/share/fonts/X11/75dpi
Login node SSH RSA/ECDSA/ED25519 host key fingerprints
The RSA, ECDSA and ED25519 fingerprints for Stanage’s login nodes are:
SHA256:mFfJmZHH0SUogoUhTtlatoZLEacfGAlj0cTrnInO5z0 (RSA)
SHA256:4HdvK3C1KDm+JG1TzxQKxezMz5ojEORynHUqF9tQfoI (ECDSA)
SHA256:aaTv+0TEc0nj7WR2ZuBYWFDD+QqzOKJpMjEFKBx6pQU (ED25519)
I have a new account, how do I transfer data from my old account
Please note that the below guide assumes that both accounts are still be active. If you have lost access to the old account in the last few weeks then get in touch with us via research-it@sheffield.ac.uk and we may be able to help transfer files across.
To transfer data between your old account and your new account you could make use of either SCP or rsync. We encourage users to use rsync as it preserves timestamps and permisions. Follow the following workflow to carry out the transfer.
Log into your new username in the cluster you want to copy to and create a folder named “OldUserAccount”.
mkdir OldUserAccount
Log into your old account and run the rsync command.
If you want to copy the files to your new account on the same cluster node (e.g., old account on Stanage to new account on Stanage), here we are only going to use the “avP” options as we dont need to compress the data.
rsync -avP /Path/To/File_Or_Directory $Your_New_UserName@$HOSTNAME:/home/$Your_New_UserName/OldUserAccount
Issue when running multiple MPI jobs in sequence
If you have multiple mpirun commands in a single batch job submission script,
you may find that one or more of these may fail after
complaining about not being able to communicate with the orted daemon on other nodes.
This appears to be something to do with multiple mpirun commands being called quickly in succession,
and connections not being pulled down and new connections established quickly enough.
Putting a sleep of e.g. 5s between mpirun commands seems to help here. i.e.
mpirun program1
sleep 5s
mpirun program2
Using ‘sudo’ or package managers to install software on the clusters
Users do not have sufficient access privileges to use sudo for any purpose. Users are not permitted to install software to the base environment
with package managers on any HPC cluster i.e. with apt-get, aptitude , zypper, emerge, pacman, yum, dnf etc…
The ability to do so will never be granted to non-system administrators because:
We need to protect the integrity of the HPC systems, e.g. the operating systems, user data and user accounts etc… Permitting the usage of sudo would allow any user to arbitrarily perform any action on the HPC system.
We need to protect the integrity of user shell environments by keeping the base HPC shell environment as bare as possible. Users cannot be permitted to install software to the base environment of the clusters as this would override and potentially pollute other user’s shell environments, break other user’s jobs and/or functionality of the entire cluster.
You are permitted to software in two places:
$HOME— personal installs that only you need; suitable for smaller packages./mnt/parscratch— for larger installs, and also suitable if you want to share with others.
You can make installed software available either just for yourself (via your .bashrc or your own module files) or for other users (via shared modules).
See Installing Applications on Stanage for guidance on installing without sudo or system package managers.
Is data encrypted at rest on HPC storage areas?
At present, no HPC storage areas on any of our clusters encrypt data at rest.
Are the HPC clusters certified to standards such as Cyber Essentials, Cyber Essentials Plus or ISO 27001?
Due to the complexity of the multi-user High Performance Computing service, the service is not currently certified as being compliant with the Cyber Essentials, Cyber Essentials Plus or ISO 27001 schemes/standards. This is unlikely to change in future.
Can I use VSCode on the HPC clusters?
Usage restrictions
Caution
The usage of VSCode on the Sheffield HPC clusters is partially restricted. Usage of the Visual Studio Code Remote - SSH and Visual Studio Code Remote Explorer extensions to run VSCode on the HPC clusters is not permitted.
The Visual Studio Code Remote - SSH and Visual Studio Code Remote Explorer extensions use SSH to download a copy of VSCode to the cluster then start VSCode on the login nodes and forward back the interface to you. This means the VSCode and all dependent processes you run in the terminal are run on the login nodes. Not only does this tend to spawn lots of processes (which might hit our 100 processes per user limit on the login nodes which will lock you out of the cluster) it also fails to clean up processes correctly when the SSH connection is eventually terminated. This results in orphaned processes using high CPU, wasting resources. Furthermore, some users also try to use large amounts of CPU by running code / debugging on the login nodes which unfairly impacts other users as well.
Hint
As documented elsewhere in this site, if you are doing anything that will require a lot of CPU or memory you should use a worker node.
Permitted alternative methods for running VSCode are detailed below in the ideal order of preference
In the first instance, we recommend a workflow where version control with Github (or similar) is used alongside VSCode where scripts/code are synchronised between machines (e.g. your local machine and the HPC cluster) using conventional Git sync commands. Users are free to use the VSCode terminal on the local machine to SSH to the clusters and execute commands where necessary.
If this is not possible then VSCode can be ran on a worker node and forwarded back to your local machine in a web browser via our VSCode Remote HPC script, (from Github). Details for its use are included on the linked Github page.
If neither of these options are feasible, then running VSCode on a local machine in concert with an SSHFS mount of the desired folders from the HPC clusters to the local filesystem is possible but discouraged due to the likelihood of poor performance from machines remote from the clusters. By mounting the folder from the HPC cluster to a local filesystem folder, users can edit files on the cluster with VSCode as if they were normal local machine files.
Launching MPI tasks with srun versus mpirun or mpiexec
Documentation found elsewhere may recommend launching MPI tasks from batch jobs
using the mpirun (or mpiexec) program that comes with the MPI implementation you are using.
For those more familiar with the use of mpirun and mpiexec, you can consider
srun to be functionally equivalent
although it takes different arguments and can also be used for starting interactive sessions on Slurm clusters.
On Stanage we recommend using srun so long as the MPI implementation is built
against a compatible version of SLURM’s PMI2 or PMIx libraries — this is true for centrally installed
OpenMPI and Intel MPI modules on Stanage.
Always use --export=ALL with srun
In batch scripts, include:
srun --export=ALL my_program
This ensures your current shell environment (including module variables) is preserved. Without it, applications may fail with errors.
Take, for instance, if we were to submit this OpenMPI non-interactive hello world job without the --export=ALL option, i.e:
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=8
module load OpenMPI/4.1.4-GCC-12.2.0
srun hello
We would encounter an error message containing:
PMIX ERROR: NOT-FOUND in file client/pmix_client.c at line 562
If using mpirun
If you must use mpirun, also set:
export SLURM_EXPORT_ENV=ALL
mpirun -np $SLURM_NTASKS my_program
Without this, mpirun may fail to find internal commands like orted, particularly on multi-node jobs, producing cryptic errors like:
An ORTE daemon has unexpectedly failed after launch...
To see what’s going wrong, add:
mpirun --mca plm_base_verbose 10 ...
This increases debug output showing how mpirun is attempting to launch processes.
Why does this matter? (PMI2/PMIx and SLURM internals)
SLURM uses PMI2 or PMIx for launching and managing MPI processes. When you load an MPI module, it often sets variables like:
SLURM_MPI_TYPE=pmix_v4
But srun (and mpirun if it internally calls srun) starts a clean environment by default.
If these variables aren’t exported (e.g. via --export=ALL or SLURM_EXPORT_ENV=ALL),
your job may break before your program even starts — hence the confusing errors.
Are there any license restrictions for ANSYS?
You may open as many ANSYS applications as you like. A per-user limit of 400 concurrent cores above the first 4 per application is enforced via a maximum of 400 multi-core licence check-outs.
Multi-core licences are:
Only required if you use more than 4 cores per application
Checked out per application
Equal to (cores used – 4)
For example, if you run one applications using 4 cores you consume 0 licenses or if you run two applications each using 12 cores you consume 16 licenses.
The 400 concurrent cores limit applies per user across all devices at once — HPC clusters, personal machines, and managed desktops all count towards the same limit.
User using ANSYS |
Multi-core licenses in use |
|---|---|
On a desktop open using 4 cores |
4 - 4 = 0 |
On another desktop using 6 cores |
6 - 4 = 2 |
A job on Stanage using 20 cores |
20 - 4 = 16 |
Another job on Stanage using 30 cores |
30 - 4 = 26 |
Total |
44 |
How can I stay connected to the cluster for longer?
Reuse SSH connections
Many SSH clients can reuse existing SSH sessions for new connections without the need to reconnect. Some clients also allow sessions to persist temporarily after you have closed all your terminal windows to allow you to easily reconnect for a short time without having to reauthenticate.
SSH clients
SSH has a built in functionality to reuse existing connections for new sessions. You can enable this feature by adding the following config to your ~/.ssh/config file on your local PC:
Host stanage.shef.ac.uk
ServerAliveInterval 30
ServerAliveCountMax 4
ControlMaster auto
ControlPath ~/.ssh/sockets/%r@%h-%p
ControlPersist 600
You will need to create the directory ~/.ssh/sockets before running ssh. The ControlPersist option allows you to specify how long (in seconds) your SSH connection
should persist after you have closed all your existing sessions. During this time you can start a new session without re-authenticating.
Danger
If you configure your SSH client to maintain connections ensure that your client PC is kept locked whenever you leave it unattended.
Warning
If you are temporarily disconnected from the network you may find that your SSH session does not immediately detect the failure. You can delete the control socket created in ~/.ssh/sockets in order to clear the session and reconnect. You should not use this option when running SSH commands on remote systems.
Putty
You can configure Putty to Share SSH connections if possible via the SSH option in the Connection Category when configuring a new connection.
As long as your existing connection remains active you can start new sessions without re-authenticating by using Duplicate Session command to start new sessions.
Other applications which use Putty for SSH connections can also re-use your existing connection without needing to reauthenticate.
Warning
If you perform a large file transfer over a shared session you may find that other sessions sharing the same connection become less responsive.
“Out of Memory”, “OOM” errors and job prematurely stopping
When “Out of Memory” (OOM) errors occur in an interactive or batch session, it indicates insufficient memory has been allocated for the job to run to completion.
See seff and sacct commands for details on memory usage/efficiency for historical or currently running jobs.
Note
When an Out-of-Memory (OOM) error occurs in a system, the metrics shown by Slurm may not be truly accurate due to the metric polling interval for Slurm being slower than the CGroup limit enforcement. This means not enough memory was given despite memory allocated being higher than the reported memory peak for the job.
Requesting higher memory normally fixes this error. See Memory Allocation for details.
How to change the ownership of files and folders when not the root user?
For security reasons only system administrators are granted access to the root account (superuser privileges) and as successfully using the chown command requires root account permissions it is not possible for a non-root user to directly reassign ownership. However, it is possible to do so indirectly by using Access Control Lists (ACLs).
In the following instructions, we will bypass these limitations by giving the second user read permissions on the data so that they can make a copy of their own, then the original user can delete the original data. It assumes user1 is the current owner and user2 is going to be the new owner:
user1 makes sure user2 has the access to the files/folders:
The files/folders have to be stored in public Fastdata areas, detailed instructions are contained in the Fastdata areas.
user1 checks the original permissions of the files/folders:
[user1@login1 [stanage] public]$ getfacl the/directory/changing/ownership/
# file: the/directory/changing/ownership/
# owner: user1
# group: clusterusers
user::rwx
group::r-x
other::r-x
Note
Before making changes to ACLs it is advisable to backup the current ACL settings using getfacl. This enables restoration in case of mistakes.
getfacl -R /full/path/to/directory > acl_backup.txt
# To reapply backed-up ACLs:
setfacl --restore=acl_backup.txt
user1 makes the files/folders available to read by user2 with Linux ACLs:
setfacl --recursive --modify u:user2:r-x the/directory/changing/ownership/
user1 ensures user2 has the access to the files/folders:
[user1@login1 [stanage] public]$ ls -l
total 4
drwxrwxr-x+ 2 user1 clusterusers 4096 Apr 3 13:52 the/directory/changing/ownership/
[user1@login1 [stanage] public]$ getfacl the/directory/changing/ownership/
# file: the/directory/changing/ownership/
# owner: user1
# group: clusterusers
user::rwx
user:user2:r-x
group::r-x
mask::r-x
other::r-x
user2 creates a temporary directory to store the files:
mkdir my/tmp/directory
user2 copies the files from user1:
cp -R /mnt/parscratch/users/user1/public/the/directory/changing/ownership/ my/tmp/directory
user2 checks if they have the copy of the files/folders with the correct ownership:
[user2@login1 [stanage] public]$ ls -l
total 4
drwxr-xr-x 2 user2 clusterusers 4096 Apr 3 14:00 the/directory/changing/ownership
Warning
user1 should check with user2 and ensure the files have been transferred prior to deletion since the fastdata areas do not have backups.
user1 deletes the existing folder recursively:
rm -rf /the/directory/changing/ownership
How do I avoid large Conda environments filling up my home directory?
Home directories have limited space and can often reach their quota limit. Conda environments exponentionally take up space, if you have or want to create one or more large Conda environments (e.g. containing bulky Deep Learning packages such as TensorFlow or PyTorch) then there’s a risk you’ll quickly use up your home directory’s storage quota.
To avoid this, build your conda environments in a fastdata area
Create a
.condarcfile in your home directory if it does not already exist.Add an
envs_dirs:andpkgs_dirs:section to your.condarcfile as shown below:
pkgs_dirs:
- /mnt/parscratch/users/$USER/anaconda/.pkg-cache/
envs_dirs:
- /mnt/parscratch/users/$USER/anaconda/.envs
We recommend users create their own personal folder in the /fastdata area. As this doesn’t exist by default, you can create it with safe permissions. See fastdata area
Then create
.envsand.pkg-cachedirectories in your fastdata area as shown below:
mkdir -p /mnt/parscratch/users/$USER/anaconda/.pkg-cache/ /mnt/parscratch/users/$USER/anaconda/.envs
Installations of environments and package caching should now occur in your fastdata area
How do I remove conda environments from my home directory?
If you are rebuilding conda environments in your fastdata area and want to remove current conda environments from your home directory follow the instructions below:
Hint
Deactivate your conda environments before removing them using source deactivate
To list your environments run:
conda info --envs
Remove environments using the following command. Replace
<environment_name>with the name of the environment you want to remove.
conda remove -n <environment_name> --all
Can I use X11 Forwarding?
Instead of X11 forwarding, Stanage uses Flight for interactive graphical sessions. Flight is based on VNC and as such is faster than X11 forwarding and has better hardware acceleration. X11 forwarding is is slow, high-latency, bandwidth intensive, and as such is no longer recommended. Flight is the preferred method for graphical HPC access.
Why is the version of glibc on Stanage still 2.17, and what should I do if I need a newer version?
If you’re getting errors like:
GLIBC_2.28 not found
then your software was likely built on a newer system and is incompatible with Stanage’s old glibc.
Stanage currently runs CentOS 7, which ships with glibc 2.17. This will not be updated until the planned OS upgrade later this year. In the meantime, if your software requires a newer glibc (e.g. 2.28+), we recommend using Apptainer containers. You can build containers based on newer distributions (e.g. Rocky 9) that provide a more modern glibc.
See our Apptainer guide for how to get started.